astropyのpyfitsを使ってfit(s)画像を読み込んで、読み込んだ画像をpycudaを使って加工して少しでも処理を高速化するというプログラムを試してみた。
スポンサーリンク
fitデータをダウンロードして加工する¶
%download http://data.astropy.org/tutorials/FITS-images/HorseHead.fits
次にダウンロードしたfits画像データをastropyのpyfitsで読み込んで加工する。
import astropy.io.fits as pyfits # Library to read FITS
import pycuda.autoinit # PyCuda autoinit
import pycuda.driver as cuda # PyCuda In, Out helpers
import matplotlib.pyplot as plot # Library to plot
import matplotlib.cm as colormap # Library to plot
import numpy # Fast math library
import time
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
pylab.rcParams['figure.figsize'] = 20, 30
pylab.rcParams["font.size"] = "20"
from pycuda.compiler import SourceModule
# Open FITS image
fits = pyfits.open('HorseHead.fits')
# Get the image data as array
data = fits[0].data.astype(numpy.float32)
# Read image metadata
header = fits[0].header
# Show all metadata
print(header.keys)
# PyCuda: Create kernel
kernel = SourceModule("""
__global__ void log_fast(float *result, float *source) {
const int i = blockIdx.x * blockDim.x + threadIdx.x;
result[i] = log10(source[i]);
}
""")
# PyCuda: Get function
log_fast = kernel.get_function("log_fast")
# PyCuda: Calculate block size
BLOCK_SIZE = 1024
block = (BLOCK_SIZE, 1, 1)
grid = (int(data.shape[0] * data.shape[1] / BLOCK_SIZE) + 1, 1, 1)
# PyCuda: Call the function to transform image
data_result = numpy.zeros_like(data)
start_time = time.time()
log_fast(cuda.Out(data_result), cuda.In(data), block=block, grid=grid)
end_time = time.time()
cuda_time = end_time - start_time
print("PyCuda log10 {0:.4f} seconds".format(cuda_time))
# NumPy
start_time = time.time()
np_data_result = numpy.log10(data)
end_time = time.time()
numpy_time = end_time - start_time
print("NumPy log10 {0:.4f} seconds ".format(numpy_time))
print("PyCuda is {0:.1f} times faster than NumPy".format(numpy_time / cuda_time))
# Show image using matplotlib
figure = plot.figure()
figure.add_subplot(3,1,1)
plot.imshow(data, cmap = colormap.Greys_r)
plt.colorbar()
figure.add_subplot(3,1,2)
plot.imshow(data_result, cmap = colormap.Greys_r)
plt.colorbar()
figure.add_subplot(3,1,3)
plot.imshow(np_data_result, cmap = colormap.Greys_r)
plt.colorbar()
plot.show()
pycudaを使うことでnumpyよりも5.5倍も画像処理速度が高速化されている。
スポンサーリンク
別の画像データを試してみる¶
別のfits画像データを下記のサイトからダウンロードする。
%download https://astropy.stsci.edu/data/photometry/spitzer_example_image.fits
ダウンロードしたfitsデータを読み込んで再びlog10画像処理する。
import astropy.io.fits as pyfits # Library to read FITS
import pycuda.autoinit # PyCuda autoinit
import pycuda.driver as cuda # PyCuda In, Out helpers
import matplotlib.pyplot as plot # Library to plot
import matplotlib.cm as colormap # Library to plot
import numpy # Fast math library
import time
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
pylab.rcParams['figure.figsize'] = 20, 30
pylab.rcParams["font.size"] = "20"
from pycuda.compiler import SourceModule
# Open FITS image
fits = pyfits.open('spitzer_example_image.fits')
# Get the image data as array
data = fits[0].data.astype(numpy.float32)
# Read image metadata
header = fits[0].header
# Show all metadata
print(header.keys)
# PyCuda: Create kernel
kernel = SourceModule("""
__global__ void log_fast(float *result, float *source) {
const int i = blockIdx.x * blockDim.x + threadIdx.x;
result[i] = log10(source[i]);
}
""")
# PyCuda: Get function
log_fast = kernel.get_function("log_fast")
# PyCuda: Calculate block size
BLOCK_SIZE = 1024
block = (BLOCK_SIZE, 1, 1)
grid = (int(data.shape[0] * data.shape[1] / BLOCK_SIZE) + 1, 1, 1)
# PyCuda: Call the function to transform image
data_result = numpy.zeros_like(data)
start_time = time.time()
log_fast(cuda.Out(data_result), cuda.In(data), block=block, grid=grid)
end_time = time.time()
cuda_time = end_time - start_time
print("PyCuda log10 {0:.4f} seconds".format(cuda_time))
# NumPy
start_time = time.time()
np_data_result = numpy.log10(data)
end_time = time.time()
numpy_time = end_time - start_time
print("NumPy log10 {0:.4f} seconds ".format(numpy_time))
print("PyCuda is {0:.1f} times faster than NumPy".format(numpy_time / cuda_time))
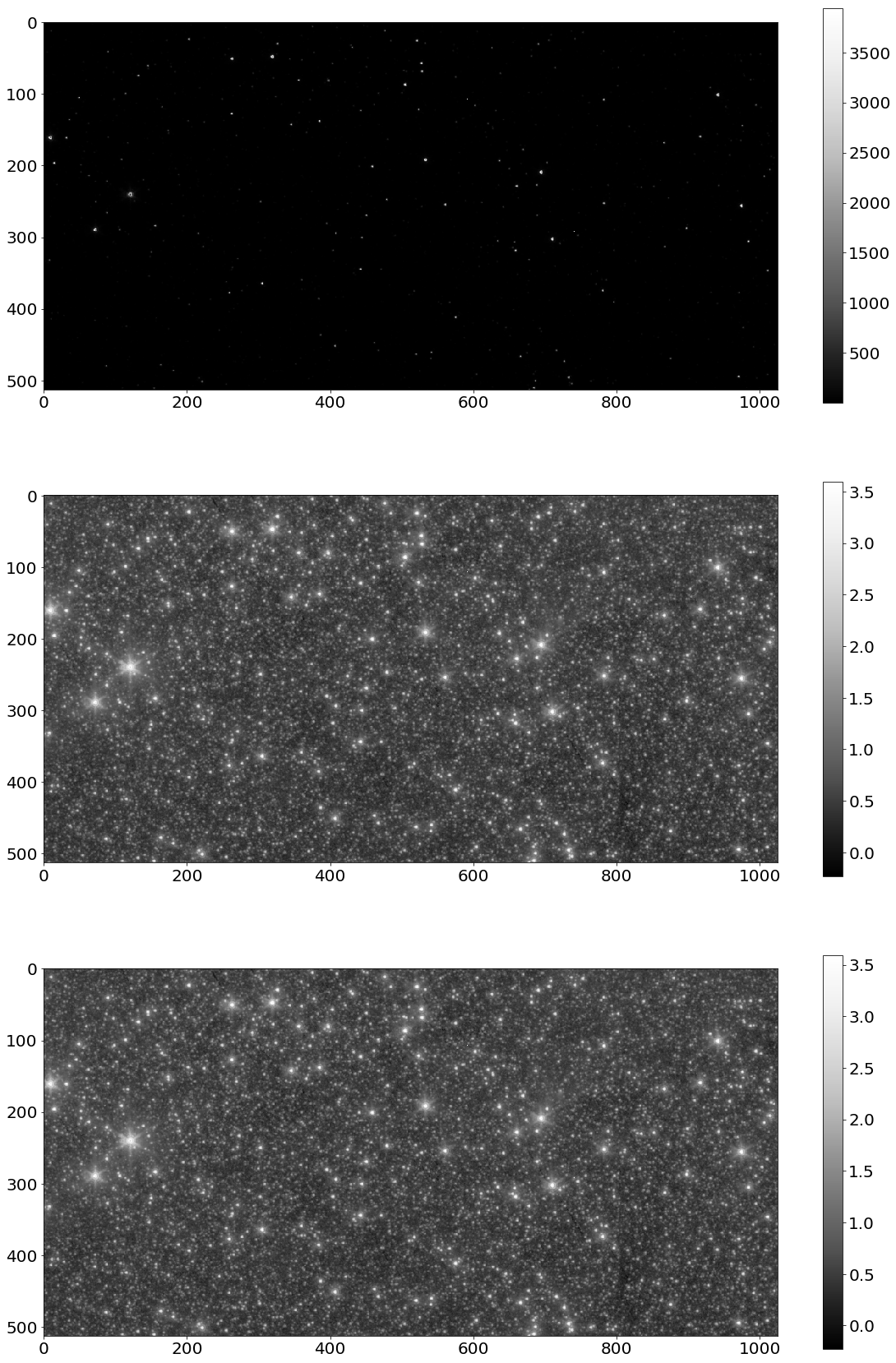
# Show image using matplotlib
figure = plot.figure()
figure.add_subplot(3,1,1)
plot.imshow(data, cmap = colormap.Greys_r)
plt.colorbar()
figure.add_subplot(3,1,2)
plot.imshow(data_result, cmap = colormap.Greys_r)
plt.colorbar()
figure.add_subplot(3,1,3)
plot.imshow(np_data_result, cmap = colormap.Greys_r)
plt.colorbar()
plot.show()
今回もpycudaを使うことでnumpyよりも処理速度が1.9倍高速化されている。ただ、色々なデータを試してみたところ、pycudaを使うことで処理速度が著しく遅くなるケースもあるので、pycudaを使用すれば必ず処理速度が上がるというわけではなくてデータのサイズによるところが大きい。
スポンサーリンク